#!pip install ANNarchySTDP - network
A simple model showing the STDP learning rule on inputs converginf to a single neuron. Model adapted from Song, Miller and Abbott (2000) and Song and Abbott (2001)
Code adapted from the Brian example: http://brian.readthedocs.org/en/1.4.1/examples-plasticity_STDP1.html
import numpy as np
import matplotlib.pyplot as plt
import ANNarchy as annANNarchy 5.0 (5.0.0) on linux (posix).Some parameters:
F = 15.0 # Poisson distribution at 15 Hz
N = 1000 # 1000 Poisson inputs
gmax = 0.01 # Maximum weight
duration = 100000.0 # Simulation for 100 secondsIntegrate-and-fire neuron:
IF = ann.Neuron(
parameters = dict(
tau_m = 10.0,
tau_e = 5.0 ,
vt = -54.0 ,
vr = -60.0 ,
El = -74.0 ,
Ee = 0.0 ,
),
equations = [
ann.Variable('tau_m * dv/dt = El - v + g_exc * (Ee - vr)', init = -60.0),
ann.Variable('tau_e * dg_exc/dt = - g_exc'),
],
spike = "v > vt",
reset = "v = vr",
)An input population of Poisson neurons, and a single post-synaptic neuron.
# Network
net = ann.Network()
# Input population
Input = net.create(ann.PoissonPopulation(geometry=N, rates=F, name = 'Input'))
# Output neuron
Output = net.create(geometry=1, neuron=IF, name = 'Output')
# Projection learned using STDP
proj = net.connect(
pre = Input,
post = Output,
target = 'exc',
synapse = ann.STDP(tau_plus=20.0, tau_minus=20.0, A_plus=0.01, A_minus=0.0105, w_max=0.01)
)
proj.all_to_all(weights=ann.Uniform(0.0, gmax))
# Compile the network
net.compile()Compiling network 1... OK # Start recording
Mi = net.monitor(Input, 'spike')
Mo = net.monitor(Output, 'spike')
# Start the simulation
net.simulate(duration, measure_time=True)
# Retrieve the recordings
input_spikes = Mi.get('spike')
output_spikes = Mo.get('spike')Simulating 100.0 seconds of the network 1 took 0.9769761562347412 seconds. # Compute the mean firing rates during the simulation
print('Mean firing rate in the input population: ' + str(Mi.mean_fr(input_spikes)) )
print('Mean firing rate of the output neuron: ' + str(Mo.mean_fr(output_spikes)) )
# Compute the instantaneous firing rate of the output neuron
output_rate = Mo.smoothed_rate(output_spikes, 100.0)
# Receptive field after simulation
weights = proj.w[0]Mean firing rate in the input population: 15.005540000000002
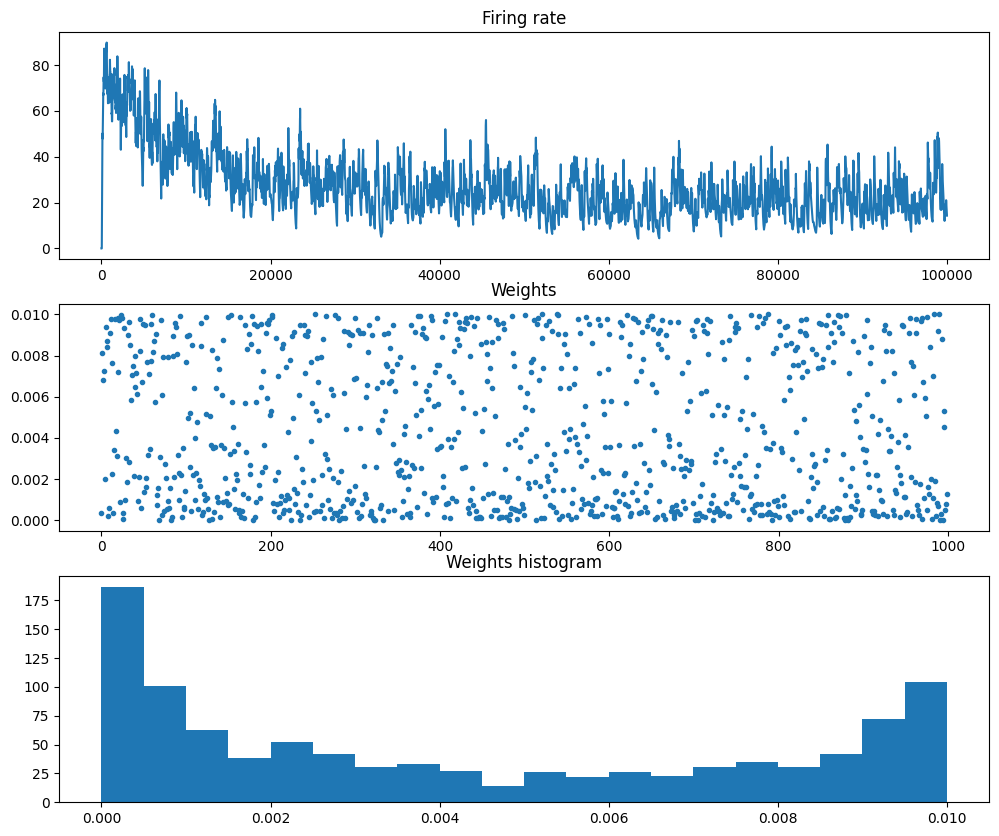
Mean firing rate of the output neuron: 29.37plt.figure(figsize=(12, 12))
plt.subplot(3,1,1)
plt.title('Firing rate')
plt.plot(output_rate[0, :])
plt.subplot(3,1,2)
plt.title('Weights')
plt.plot(weights, '.')
plt.subplot(3,1,3)
plt.title('Weights histogram')
plt.hist(weights, bins=20)
plt.show()