#!pip install ANNarchyGap Junctions
A simple network with gap junctions.
This is a reimplementation of the Brian example:
http://brian2.readthedocs.org/en/2.0b3/examples/synapses.gapjunctions.html
import numpy as np
import matplotlib.pyplot as plt
import ANNarchy as ann
neuron = ann.Neuron(
parameters = dict(v0 = 1.05, tau = 10.0),
equations = "tau * dv/dt = v0 - v + g_gap",
spike = "v > 1.",
reset = "v = 0."
)
gap_junction = ann.Synapse(
psp = "w * (pre.v - post.v)"
)
net = ann.Network(dt=0.1)
pop = net.create(10, neuron)
pop.v = np.linspace(0., 1., 10)
proj = net.connect(pop, pop, 'gap', gap_junction)
proj.all_to_all(0.02)
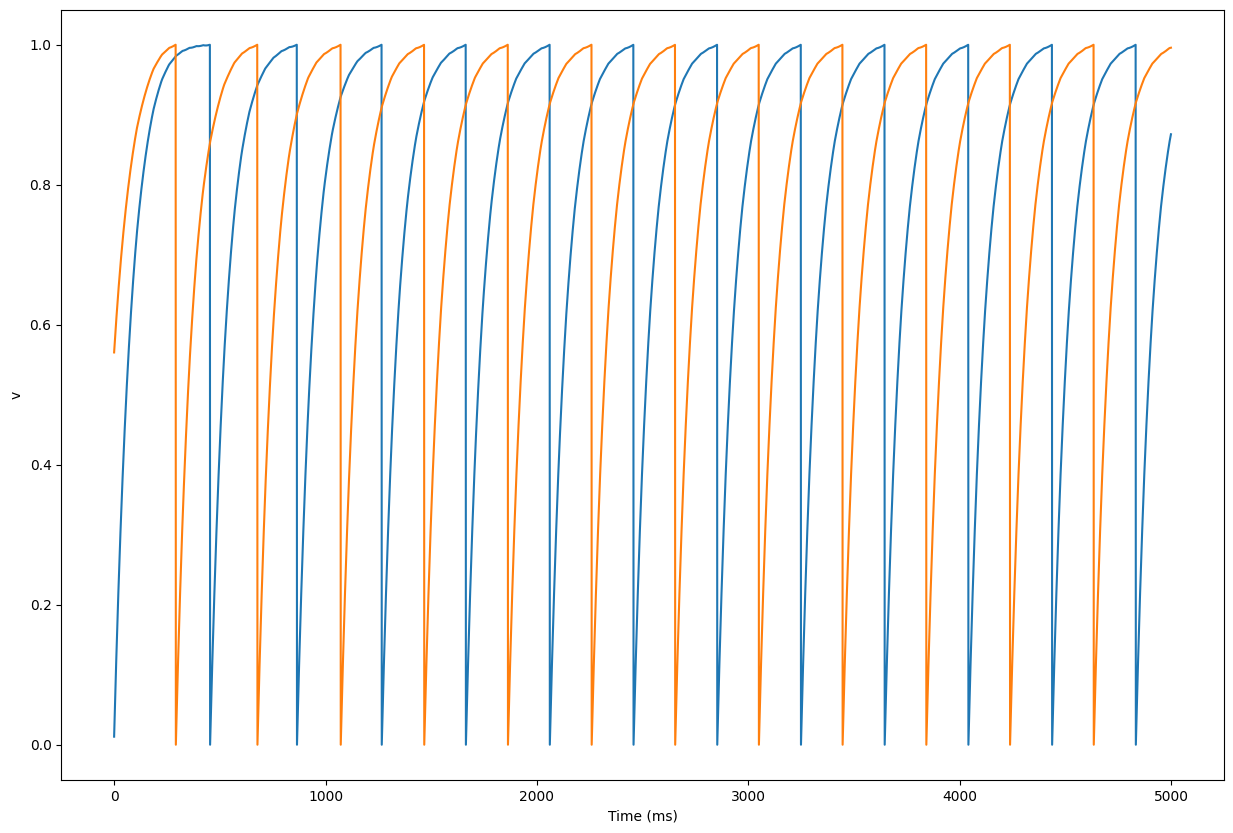
trace = net.monitor(pop[0] + pop[5], 'v')
net.compile()
net.simulate(500.)
data = trace.get('v')
plt.figure(figsize=(10, 8))
plt.plot(data[:, 0])
plt.plot(data[:, 1])
plt.xlabel('Time (ms)')
plt.ylabel('v')
plt.show()ANNarchy 5.0 (5.0.0) on linux (posix).
Compiling network 1... OK