#!pip install ANNarchyParallel simulations
This example demonstrates the use of parallel_run() to simulate the same network multiple times in parallel.
import numpy as np
import matplotlib.pyplot as plt
import ANNarchy as ann
import timeANNarchy 5.0 (5.0.0) on linux (posix).Parallel simulations use the multiprocessing module to start parallel processes. On Linux, it should work directly, but there is an issue on OSX. Since Python 3.8, the ‘spawn’ method is the default way to start processes, but it does not work on MacOS. The following cell should fix the issue, but it should only be ran once.
import platform
if platform.system() == "Darwin":
import multiprocessing as mp
mp.set_start_method('fork')We start by creating the Izhikevich pulse-coupled network defined in Izhikevich.ipynb, but using a class inheriting from ann.Network:
class PulseNetwork(ann.Network):
def __init__(self, size=1000):
self.rng = np.random.default_rng(seed=self.seed)
# Create the population
self.P = self.create(geometry=size, neuron=ann.Izhikevich)
# Create the excitatory population
nb_exc = int(0.8*size)
self.Exc = self.P[:nb_exc]
re = self.rng.random(nb_exc)
self.Exc.noise = 5.0
self.Exc.a = 0.02
self.Exc.b = 0.2
self.Exc.c = -65.0 + 15.0 * re**2
self.Exc.d = 8.0 - 6.0 * re**2
self.Exc.v = -65.0
self.Exc.u = self.Exc.v * self.Exc.b
# Create the Inh population
self.Inh = self.P[nb_exc:]
ri = self.rng.random(size - nb_exc)
self.Inh.noise = 2.0
self.Inh.a = 0.02 + 0.08 * ri
self.Inh.b = 0.25 - 0.05 * ri
self.Inh.c = -65.0
self.Inh.d = 2.0
self.Inh.v = -65.0
self.Inh.u = self.Inh.v * self.Inh.b
# Create the projections
self.proj_exc = self.connect(self.Exc, self.P, 'exc')
self.proj_inh = self.connect(self.Inh, self.P, 'inh')
self.proj_exc.all_to_all(weights=ann.Uniform(0.0, 0.5, self.rng))
self.proj_inh.all_to_all(weights=ann.Uniform(0.0, 1.0, self.rng))
# Create a spike monitor
self.M = self.monitor(self.P, 'spike')We create such a network, compile it and define a simulation method. Here, it is only a simple simulation, while the methods returns the recorded spike trains.
# Create network
net = PulseNetwork(1000)
net.config(dt=1.0)
net.compile()
# Single trial
def run(net, duration=1000):
# Simulate
net.simulate(duration)
# Recordings
t, n = net.M.raster_plot()
return t, n
t, n = run(net, 1000)
plt.figure()
plt.plot(t, n, '.')
plt.show()Compiling network 1... OK 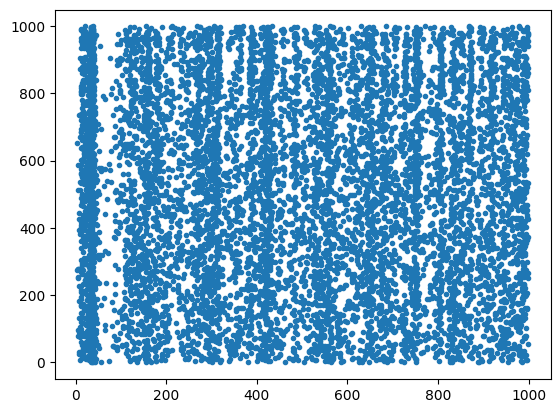
Using the parallel_run methods of Network allows to reinitialize the network and run the simulation multiple times in parallel, with different durations. The results of each simulation are concatenated and returned.
We make sure that the seeds of each network are different by setting the value 'sequential'. None or a list of seeds would also work.
Note how, in the network definition, the rng of the random distributions was set using the seed of the network.
results = net.parallel_run(
method=run,
number=8,
seeds='sequential',
duration=[200 * (i+1) for i in range(8)]
)We can then plot the 8 simulations
plt.figure(figsize=(15, 15))
for i in range(8):
t, n = results[i]
plt.subplot(4, 2, i+1)
plt.plot(t, n, '.')
plt.show()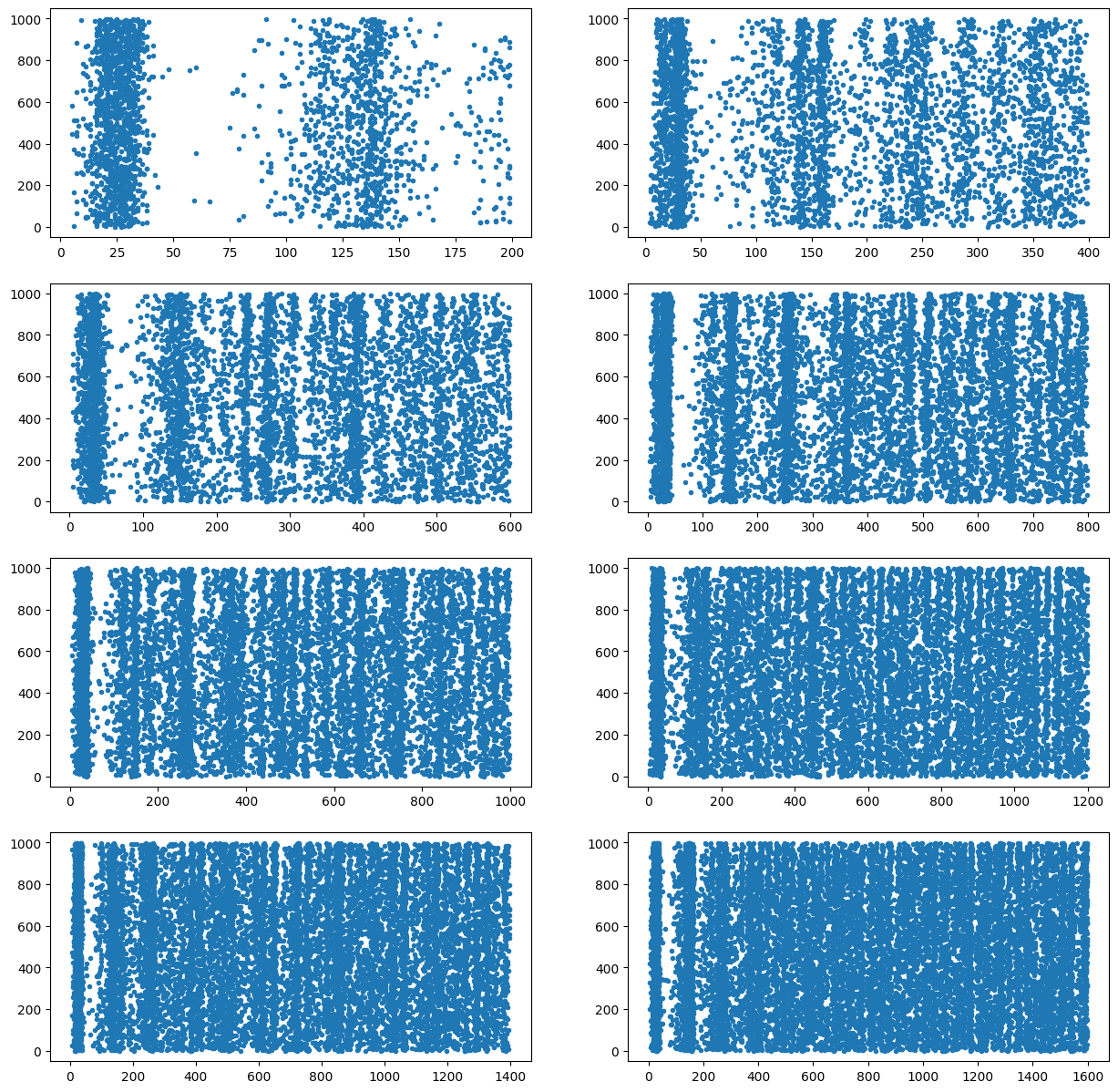
As a side note, it is important to define the network as a subclass of ann.Network, not just as an instance of ann.Network() where populations are created manually. parallel_run() needs to create copies of the network instance, which is only possible if the constructor of the class has all the required information to build those instances.
The following code should crash:
net = ann.Network()
pop = net.create(geometry=1000, neuron=ann.Izhikevich)
Exc = pop[:800]
Inh = pop[800:]
re = np.random.random(800) ; ri = np.random.random(200)
Exc.noise = 5.0 ; Inh.noise = 2.0
Exc.a = 0.02 ; Inh.a = 0.02 + 0.08 * ri
Exc.b = 0.2 ; Inh.b = 0.25 - 0.05 * ri
Exc.c = -65.0 + 15.0 * re**2 ; Inh.c = -65.0
Exc.d = 8.0 - 6.0 * re**2 ; Inh.d = 2.0
Exc.v = -65.0 ; Inh.v = -65.0
Exc.u = Exc.v * Exc.b ; Inh.u = Inh.v * Inh.b
exc_proj = net.connect(pre=Exc, post=pop, target='exc')
exc_proj.all_to_all(weights=ann.Uniform(0.0, 0.5))
inh_proj = net.connect(pre=Inh, post=pop, target='inh')
inh_proj.all_to_all(weights=ann.Uniform(0.0, 1.0))
m = net.monitor(pop, 'spike')
net.compile()
net.parallel_run(number=2, method=run)Compiling network 2... OK --------------------------------------------------------------------------- ANNarchyException Traceback (most recent call last) Cell In[8], line 26 22 m = net.monitor(pop, 'spike') 24 net.compile() ---> 26 net.parallel_run(number=2, method=run) File ~/Dokumente/git-repos/ANNarchy/ANNarchy/core/Network.py:721, in Network.parallel_run(self, method, number, max_processes, seeds, measure_time, **kwargs) 718 Messages._debug("Network was created with ", self._init_args, "and", self._init_kwargs) 720 if type(self) is Network: --> 721 Messages._error("Network.parallel_run(): the network must be an instance of a class deriving from Network, not Network itself.") 723 if measure_time: 724 tstart = time.time() File ~/Dokumente/git-repos/ANNarchy/ANNarchy/intern/Messages.py:115, in _error(*var_text, **args) 112 exit = True 114 if exit: --> 115 raise ANNarchyException(text) 116 else: 117 print('ERROR:' + text) ANNarchyException: Network.parallel_run(): the network must be an instance of a class deriving from Network, not Network itself.

